Defining the determinants of the response to therapeutic exposure in cancer
Cancer cells incorporate a large number of internal and external cues as they execute oncogenic processes. The treatment of cancer induces complex molecular changes across these programs. These are dependent on the mechanism of action of the applied therapy as well as due to differences in genetic background both between patients and within an individual tumor. Defining the regulation of these innate and treatment-induced landscapes can identify successful points of therapeutic intervention. The Chemical Genomics Laboratory at Columbia aims to leverage cutting-edge multiplex single-cell genetic and chemical genomics tools to generate these maps of cellular response.
Our approach
Multiplex single-cell chemical transcriptomics
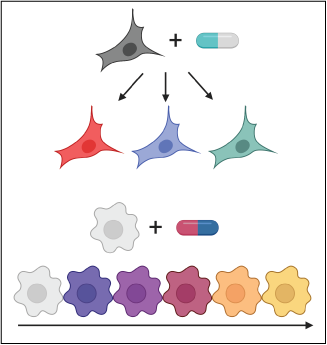
Our sci-Plex multiplexing strategy enables the coupling of high-throughput chemical screens to single-cell RNA-seq. We aim to use this platform to characterize the molecular effects of the treatment of tumor cells to thousands of unique therapeutic exposures. Our goal is to leverage these atlases to uncover cell type and cell state specific responses that present vulnerabilities or paths to resistance that can inform new therapeutic strategies.
CRISPR-based single-cell molecular screens
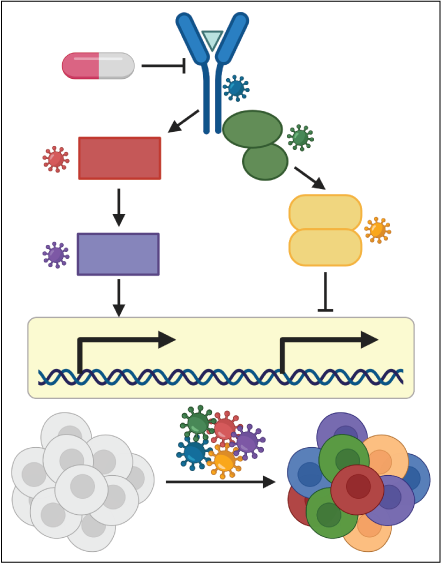
Single-cell pooled CRISPR screens allow for the molecular characterization of the effect of many genetic perturbations in parallel. Combined with chemical transcriptomics these screens enable the dissection of the contribution of individual genes to drug-induced molecular responses. We aim to use these techniques to identify the gene regulatory networks causative for individual aspects of the complex response initiated by tumor cells after therapeutic exposure.
In vivo single-cell perturbation genomics
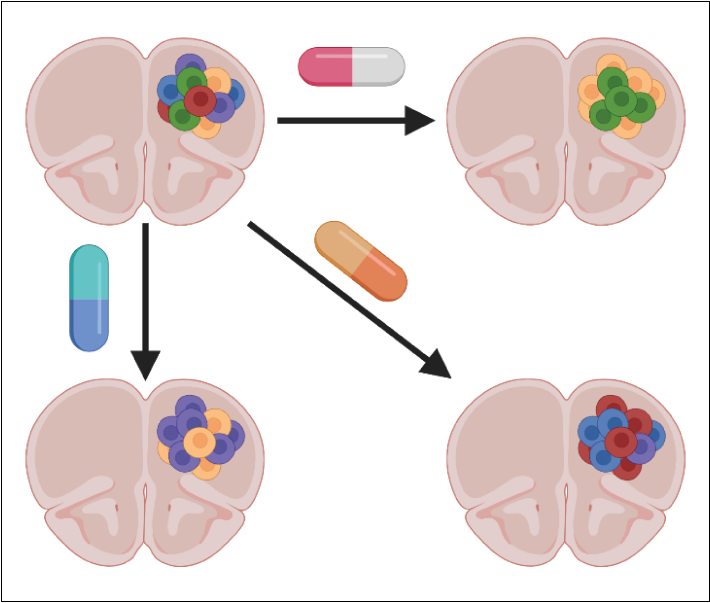
We are interested in developing and extending our tools for single-cell perturbation screens to determine how genetic and chemical perturbations interact to drive therapeutic response in tissues. Combined with our large-scale in vitro efforts, these approaches will allow us to identify the molecular changes associated with response to therapy in the context of the complex interactions between cancer cells and their environment.